Development of a human in vitro model for retinitis pigmentosa
Tissue
Retina ModelsThe Problem of the Client
The client was studying retinitis pigmentosa (RP), more specifically mutation RHO-P23H, which is the main cause for autosomal dominant RP. The phenotypes of animal models vary depending on the model, thus the client wanted to generate a human physiologically-relevant RHO-P23H in vitro model to better understand the disease mechanisms and ultimately improve clinical translation of new therapies for patients with the mutation.

Solution
We generated an iPSC cell line with the RHO-P23H mutation using CRISPRCas9 gene editing and differentiated the line into retinal organoids to analyse their cellular composition.
Step 1:
Understanding the Problem
Our technical experts defined the detailed problem and questions to be answered through in-depth discussion with the client.
It was crucial for the client to analyse the impact of a common RP mutation on the cellular composition of retinal microtissue. Therefore, the heterozygous RHO-P23H knock-in mutation had to be introduced into the RHO gene of human iPSCs derived from a healthy donor using CRISPRCas9– mediated gene editing.
It was also necessary to compare, at specific timepoints, isogenic wild type (WT) and mutated retinal organoids derived from the iPSC lines for morphology, protein expression profile and ultrastructure in order to characterise the phenotypic changes caused by the heterozygous mutation in the RHO gene.
Step 2:
Developing a Customised Experimental Plan
We designed a study in 4 parts:
- Generation of RHO-P23H iPSCs cell line.
- Differentiation of RHO-P23H and WT iPSCs into mature retinal organoids until days 180, 210 and 240 of differentiation.
- Quantitative immunofluorescence analysis of cell specific biomarkers to quantify rod photoreceptors, connecting cilia and astrocytes with a with a Zeiss Axio Apotome fluorescence microscope.
- Transmission electron microscopy (TEM) to analyse ultrastructural differences between WT and RHO-P23H retinal organoids.
Step 3:
Project Execution
The study was carried out by our team of experts, providing regular updates to the client.
The study included the experimental phase, data processing, data analysis and presentation of a comprehensive data summary.
Output Data Set:
The complete dataset derived from the study included immunofluorescent quantification data of cells from mutated and wild type organoids compared at three different timepoints of differentiation.
- Organoid lines compared: RHO-P23H, Isogenic WT control
- Time points of organoid differentiation: day 180, day 210 and day 240
- Cell types quantified (cell specific marker): rod photoreceptors (RHO), cone photoreceptor (OPN1MW/LW), connecting cilia (ARL13B) and astrocytes (GAFP).
- Number of organoids analysed per experimental condition: 6
Example data set that was provided to the client:
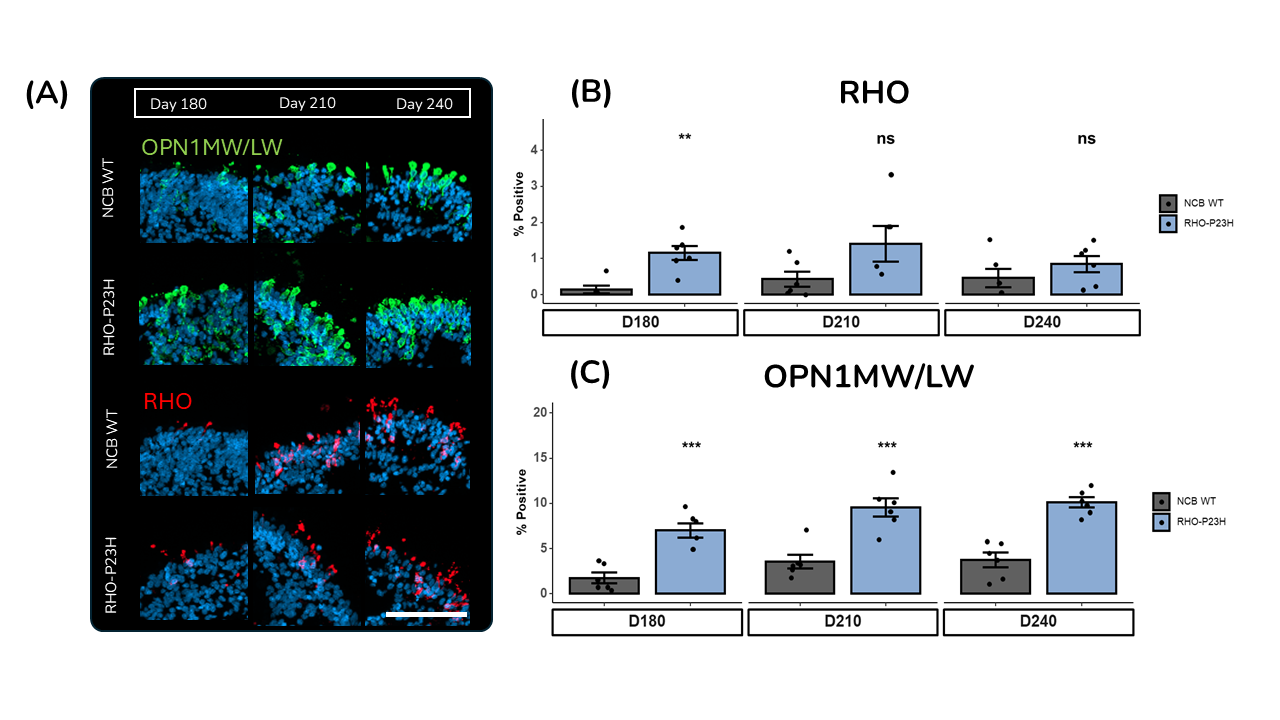
(A) Representative immunofluorescence images of RO day 180, 210 and 240 of differentiation. Scale bar = 50 μm.
(B) IF quantification of rod photoreceptors. ROs were stained, quantified for percentage positivity and analysed for differences between RHO-P23H and NCB WT ROs within each timepoint by t-test. n=6 ROs per condition. Data bars represent mean ± SD, individual points represent quantification of single ROs.
(C) IF quantification of cone photoreceptors. ROs were stained, quantified for percentage positivity and analysed for differences between RHO-P23H and NCB WT ROs within each timepoint by t-test. n=6 ROs per condition. Data bars represent mean ± SD, individual points represent quantification of single ROs.
Step 4:
Delivery of Results
We delivered a detailed report of the dataset which was shared digitally and discussed over a call.
The study highlighted:
- Ability to generate a human in vitro retinal organoids model relevant to RP.
- Significant increase of protein expression of the RHO marker for the rod photoreceptor in RHO-P23H organoids at day 180 but no longer at day 210 and 240.
- The marker for cone photoreceptors (OPN1MW/LW) increase over time from day 180 to day 240 in the RP model.
- Lower cilia marker expression (ARL13B) at day 180 in RHO-P23H organoids compared to WT, with a decrease over time.
- Reduction of astrocytes marker expression at day 180, day 210 and day 240 in the RP model compared to wild type.
- Increased signs of cellular stress identified by TEM in RP model organoids such as stress vacuoles.
Outcomes for the Client and the Project
The client obtained a robust, fully characterised and relevant in vitro human RP retinal tissue model (RHO-P23H) that can be used for further testing of novel therapeutics. The client also acquired valuable insights into the pathophysiology of RP including the impairment of cellular trafficking to the outer segment.